D Tables of plasmids and strains
D.1 Plasmid and strain tables
| Code | Description |
|---|---|
| ha3_2 | pSC101 carb pCinLacO_m B0032 CinI BCD2 sfCFP |
| har_2 | p15a chlor pCinTetO B0032 LacI B0034 mScarlet-I |
| i1f_8 | p15a chlor J23106 B0034 mScarlet-I |
| i38_13 | pSC101 carb pRhlLacO_m B0032 RhlI-ssrA BCD2 sfCFP |
| i39_1 | p15a chlor pRhl B0032 LacI-ssrA B0034 mScarlet-I-ssrA |
| j78_7 | pSC101 carb pCinLacO_m B0032 truncated-CinI-ssrA BCD2 sfCFP |
| j7h_1 | p15a chlor pCinTetO truncated-LacI mScarlet-I |
| jag_d3 | pSC101 carb pRpa B0034 CinI BCD2 sfYFP T J23103 B0032 RpaR |
| Description | Code | Plasmids | E. coli strain |
|---|---|---|---|
| Cin Amplifier strain | i59_2 | ha3_2;har_2 | CY026 |
| Weak sender strain | jao_1 | jag_d3;har_2 | CY026 |
| Strong sender strain | k25_1 | jag_d3;j78_7 | CY026 |
| Reporter strain | k25_3 | j7h_1;j78_7 | CY026 |
| Constitutive mScarlet-I | i1f_8 | i1f_8 | JS006 |
| Rhl positive feedback strain | i38_13 | i38_13 | CY026 |
| Description | Genotype | Source |
|---|---|---|
| CY026 | BW25113 ΔlacI ΔaraC ΔsdiA Ptrc* -cinR Ptrc* -rhlR | Ye Chen, Addgene Bacterial strain #72340 |
| JS006 | BW25113 ΔaraC ΔlacI | Jesse Stricker, Stricker et al 2008 |
D.2 Plasmids maps
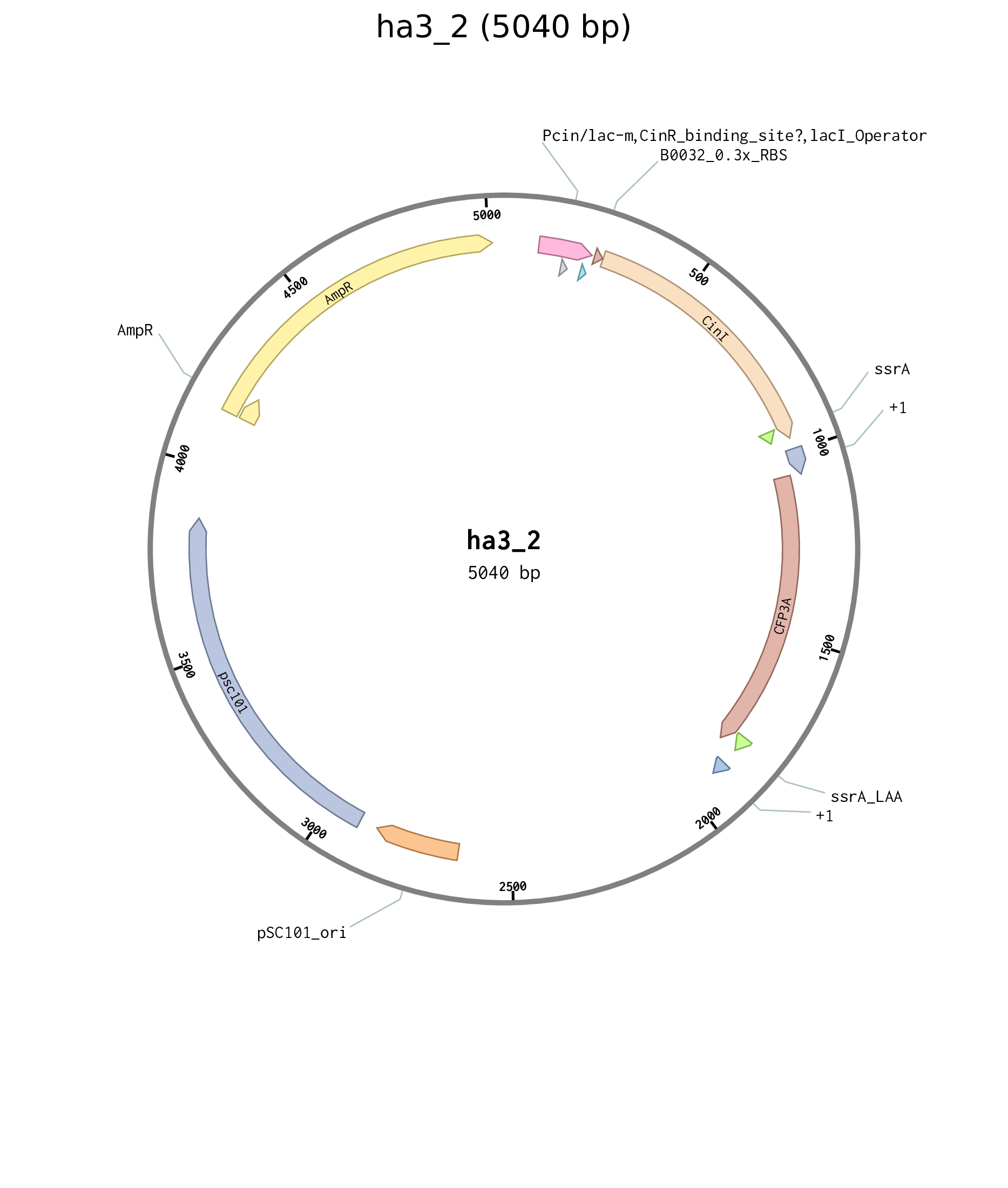
Figure D.1: Plasmid map for Cin positive feedback plasmid ha3_2.
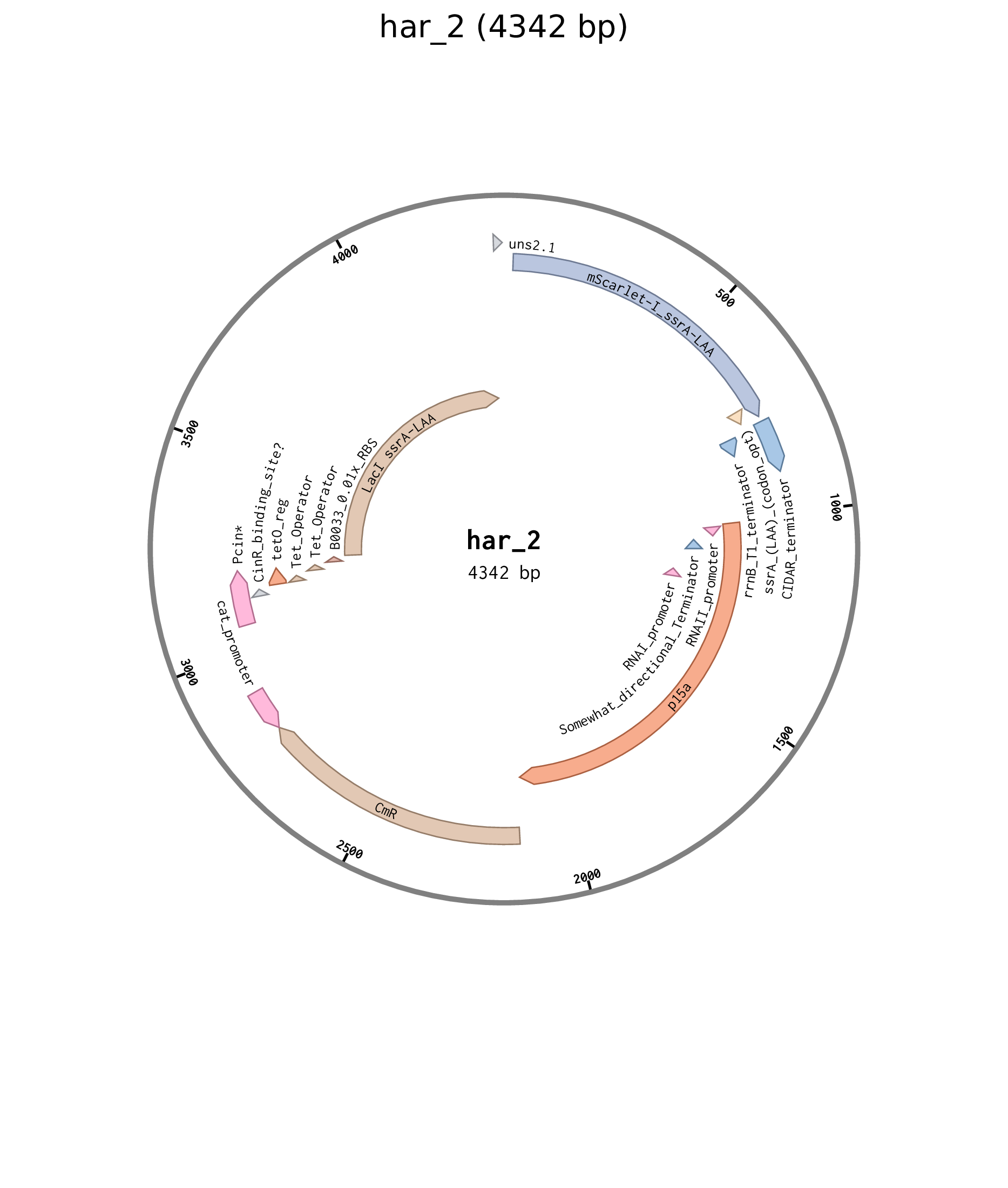
Figure D.2: Plasmid map for Cin negative feedforward plasmid har_2.

Figure D.3: Plasmid map for constitutive mScarlet-I source i1f_8.

Figure D.4: Plasmid map for Rhl positive feedback plasmid i38_13.
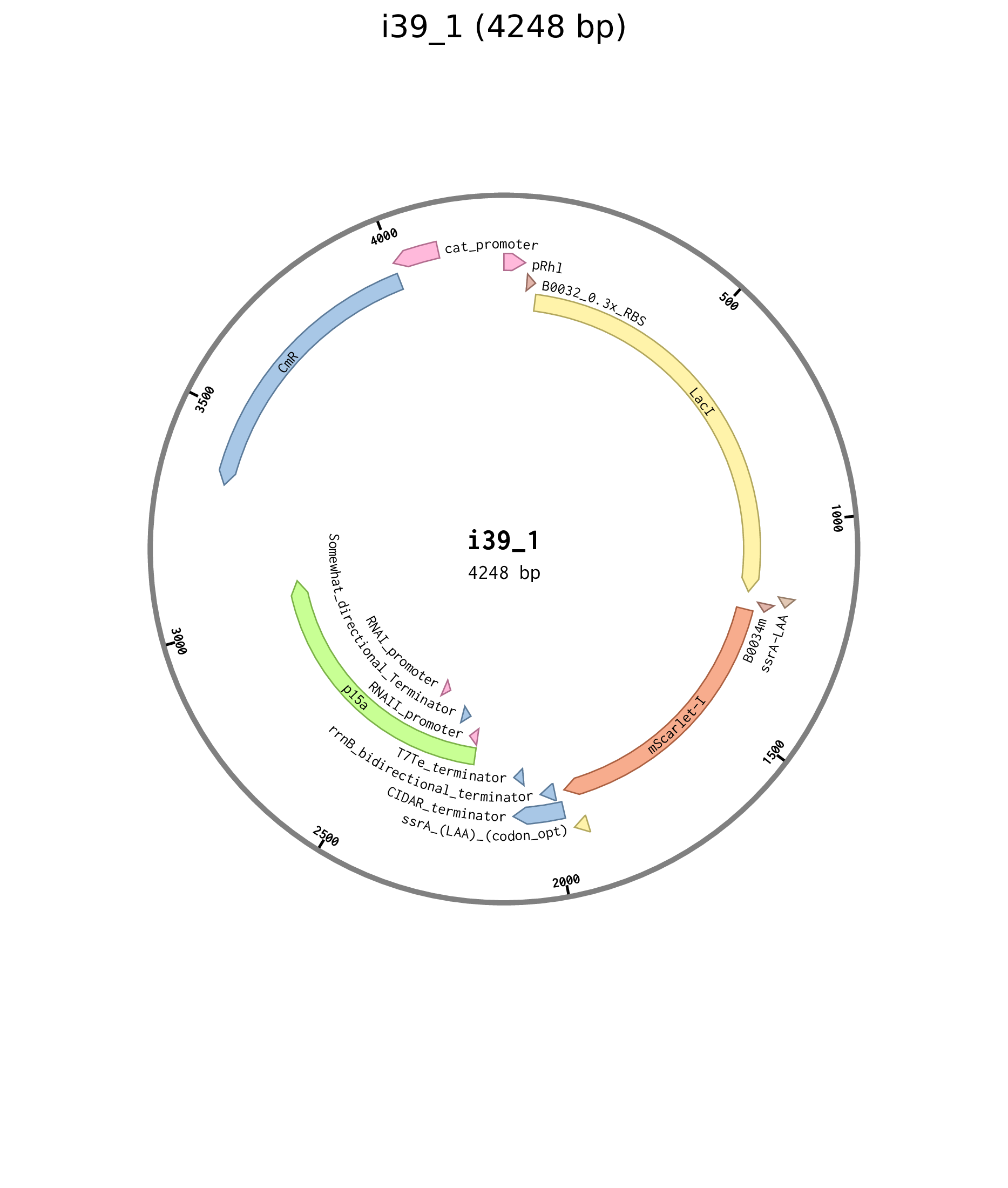
Figure D.5: Plasmid map for Rhl negative feedforward plasmid i39_1.
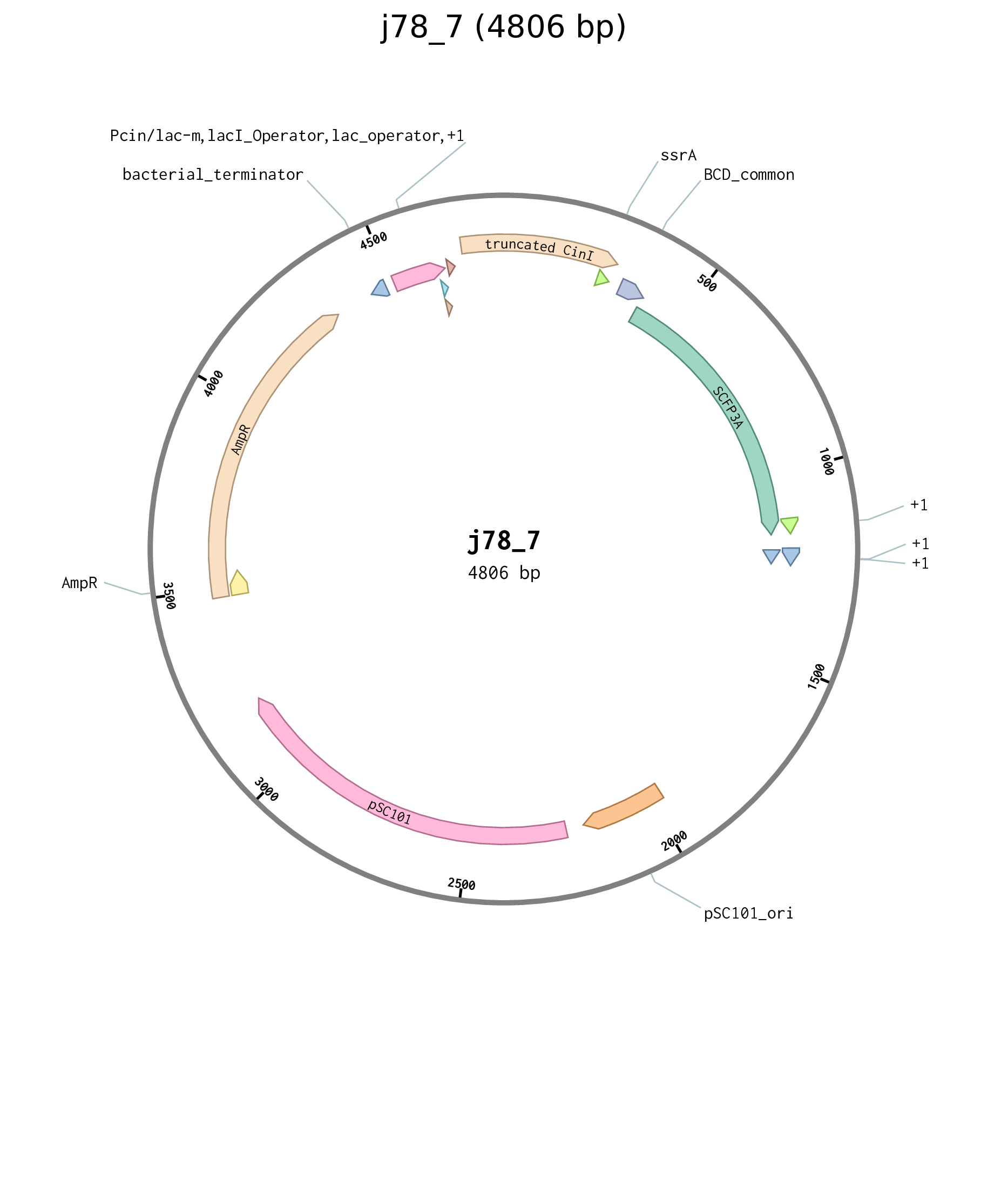
Figure D.6: Plasmid map for truncated CinI Cin reporter plasmid j78_7.
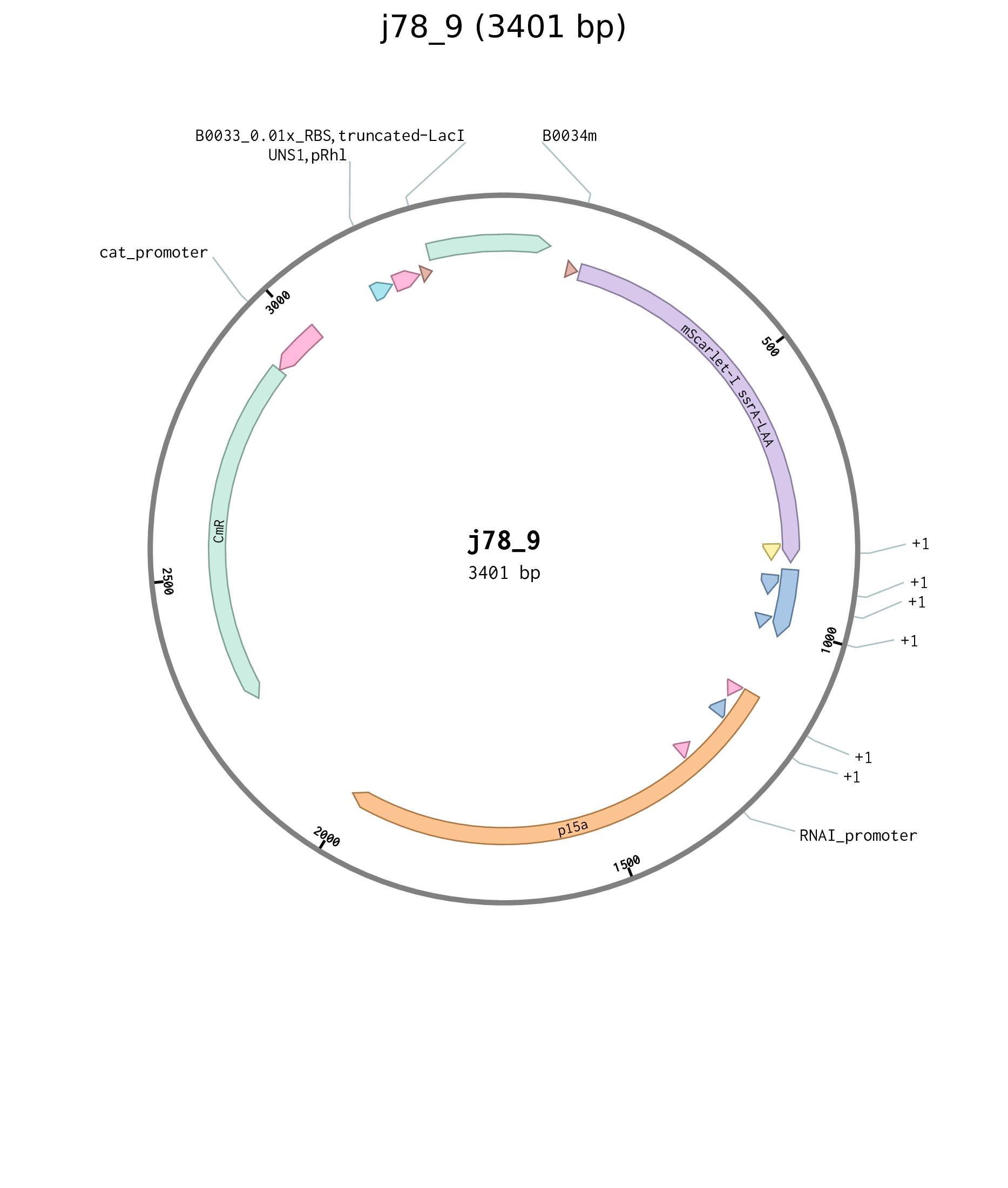
Figure D.7: Plasmid map for truncated LacI Cin reporter plasmid j78_9.

Figure D.8: Plasmid map for truncated LacI Rhl reporter plasmid j7h_1.
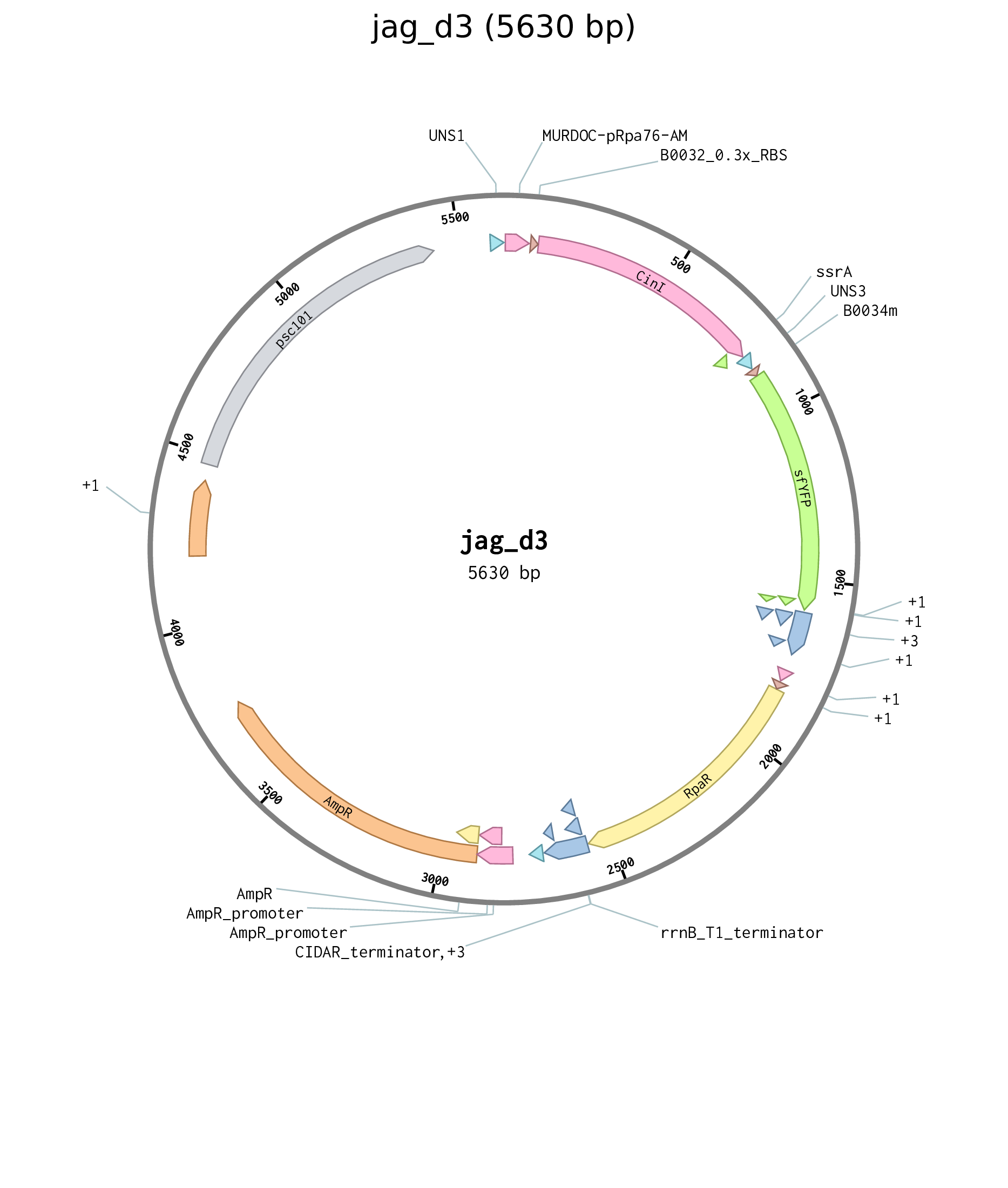
Figure D.9: Plasmid map for Cin sender plasmid jag_d3.
Alon, Uri. 2006. An introduction to systems biology: Design principles of biological circuits.
Avis, Tyler J., Valérie Gravel, Hani Antoun, and Russell J. Tweddell. 2008. “Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity.” Soil Biology and Biochemistry 40 (7): 1733–40. https://doi.org/10.1016/j.soilbio.2008.02.013.
Balagaddé, Frederick K., Hao Song, Jun Ozaki, Cynthia H. Collins, Matthew Barnet, Frances H. Arnold, Stephen R. Quake, and Lingchong You. 2008. “A synthetic Escherichia coli predator-prey ecosystem.” Molecular Systems Biology 4 (187): 187. https://doi.org/10.1038/msb.2008.24.
Balleza, Enrique, J. Mark Kim, and Philippe Cluzel. 2018. “Systematic characterization of maturation time of fluorescent proteins in living cells.” Nature Methods 15 (1): 47–51. https://doi.org/10.1038/nmeth.4509.
Basu, Subhayu, Yoram Gerchman, Cynthia H. Collins, Frances H. Arnold, and Ron Weiss. 2005. “A synthetic multicellular system for programmed pattern formation.” Nature 434 (7037): 1130–4. https://doi.org/10.1038/nature03461.
Belousov, B. P. 1959. “A periodic reaction and its mechanism.” Collection of Short Papers on Radiation Medicine.
Berlec, Aleš, Janja Završnik, Miha Butinar, Boris Turk, and Borut Štrukelj. 2015. “In vivo imaging of Lactococcus lactis, Lactobacillus plantarum and Escherichia coli expressing infrared fluorescent protein in mice.” Microbial Cell Factories. https://doi.org/10.1186/s12934-015-0376-4.
Bulgarelli, Davide, Matthias Rott, Klaus Schlaeppi, Emiel Ver Loren van Themaat, Nahal Ahmadinejad, Federica Assenza, Philipp Rauf, et al. 2012. “Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota.” Nature 488 (7409): 91–95. https://doi.org/10.1038/nature11336.
Cao, Yangxiaolu, Marc D. Ryser, Stephen Payne, Bochong Li, Christopher V. Rao, and Lingchong You. 2016. “Collective Space-Sensing Coordinates Pattern Scaling in Engineered Bacteria.” Cell 165 (3): 620–30. https://doi.org/10.1016/j.cell.2016.03.006.
Chen, Ye, Jae Kyoung Kim, Andrew J. Hirning, Krešimir Josić, and Matthew R. Bennett. 2015. “Emergent genetic oscillations in a synthetic microbial consortium.” Science 349 (6251): 986–89. https://doi.org/10.1126/science.aaa3794.
Cheng, Xianrui, and James E. Ferrell. 2018. “Apoptosis propagates through the cytoplasm as trigger waves.” Science 361 (6402): 607–12. https://doi.org/10.1126/science.aah4065.
Christian, David A., Anita A. Koshy, Morgan A. Reuter, Michael R. Betts, John C. Boothroyd, and Christopher A. Hunter. 2014. “Use of transgenic parasites and host reporters to dissect events that promote interleukin-12 production during toxoplasmosis.” Infection and Immunity 82 (10): 4056–67. https://doi.org/10.1128/IAI.01643-14.
Daeffler, Kristina N‐M, Jeffrey D Galley, Ravi U Sheth, Laura C Ortiz‐Velez, Christopher O Bibb, Noah F Shroyer, Robert A Britton, and Jeffrey J Tabor. 2017. “Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation.” Molecular Systems Biology 13 (4): 923. https://doi.org/10.15252/msb.20167416.
Dalchau, Neil, Gregory Szép, Rosa Hernansaiz-Ballesteros, Chris P. Barnes, Luca Cardelli, Andrew Phillips, and Attila Csikász-Nagy. 2018. “Computing with biological switches and clocks.” Natural Computing 17 (4): 761–79. https://doi.org/10.1007/s11047-018-9686-x.
Danino, Tal, Octavio Mondragón-Palomino, Lev Tsimring, and Jeff Hasty. 2010. “A synchronized quorum of genetic clocks.” Nature 463 (7279): 326–30. https://doi.org/10.1038/nature08753.
Dieterle, Paul B, Jiseon Min, Daniel Irimia, and Ariel Amir. 2020. “Dynamics of diffusive cell signaling relays.” eLife 9. https://doi.org/10.7554/elife.61771.
Dilanji, Gabriel E., Jessica B. Langebrake, Patrick De Leenheer, and Stephen J. Hagen. 2012. “Quorum activation at a distance: Spatiotemporal patterns of gene regulation from diffusion of an autoinducer signal.” Journal of the American Chemical Society 134 (12): 5618–26. https://doi.org/10.1021/ja211593q.
Dockery, Jack D., and James P. Keener. 2001. “A mathematical model for quorum sensing in Pseudomonas aeruginosa.” Bulletin of Mathematical Biology 63 (1): 95–116. https://doi.org/10.1006/bulm.2000.0205.
Doelman, Arjen. 2019. “Pattern formation in reaction-diffusion systems- an explicit approach.” https://doi.org/10.1142/9789813239609_0004.
Donaldson, Gregory P., S. Melanie Lee, and Sarkis K. Mazmanian. 2015. “Gut biogeography of the bacterial microbiota.” Nature Reviews Microbiology 14 (1): 20–32. https://doi.org/10.1038/nrmicro3552.
Doong, Joy, James Parkin, and Richard M. Murray. 2017. “Length and time scales of cell-cell signaling circuits in agar.” bioRxiv, 1–19. https://doi.org/10.1101/220244.
Farr, B., and W. M. Farr. 2015. “Kombine: A Kernel-Density-Based, Embarrassingly Parallel Ensemble Sampler.” In Preparation.
Foreman-Mackey, Daniel, David W. Hogg, Dustin Lang, and Jonathan Goodman. 2012. “emcee: The MCMC Hammer.” Publications of the Astronomical Society of the Pacific 125 (925): 306–12. https://doi.org/10.1086/670067.
Gantner, Stephan, Michael Schmid, Christine Dürr, Regina Schuhegger, Anette Steidle, Peter Hutzler, Christian Langebartels, Leo Eberl, Anton Hartmann, and Frank B. Dazzo. 2006. “In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots.” FEMS Microbiology Ecology 56 (2): 188–94. https://doi.org/10.1111/j.1574-6941.2005.00037.x.
Garcia-Ojalvo, Jordi, Michael B. Elowitz, and Steven H. Strogatz. 2004. “Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing.” Proceedings of the National Academy of Sciences of the United States of America 101 (30): 10955–60. https://doi.org/10.1073/pnas.0307095101.
Gelens, Lendert, Graham A. Anderson, and James E. Ferrell. 2014. “Spatial trigger waves: Positive feedback gets you a long way.” Molecular Biology of the Cell 25 (22): 3486–93. https://doi.org/10.1091/mbc.E14-08-1306.
Gines, G, A S Zadorin, J.-C Galas, T Fujii, A Estevez-Torres, and Y Rondelez. 2017. “Microscopic agents programmed by DNA circuits.” NATURE NANOTECHNOLOGY | 12. https://doi.org/10.1038/NNANO.2016.299.
Goentoro, Lea, Oren Shoval, Marc W. Kirschner, and Uri Alon. 2009. “The Incoherent Feedforward Loop Can Provide Fold-Change Detection in Gene Regulation.” Molecular Cell 36 (5): 894–99. https://doi.org/10.1016/j.molcel.2009.11.018.
Goldbeter, Albert. 2006. “Oscillations and waves of cyclic AMP in Dictyostelium: A prototype for spatio-temporal organization and pulsatile intercellular communication.” Bulletin of Mathematical Biology 68 (5): 1095–1109. https://doi.org/10.1007/s11538-006-9090-z.
Gupta, Apoorv, Irene M.Brockman Reizman, Christopher R. Reisch, and Kristala L. J. Prather. 2017. “Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit.” Nature Biotechnology 35 (3): 273–79. https://doi.org/10.1038/nbt.3796.
Gupta, Sonali, Tyler D. Ross, Marcella M. Gomez, Job L. Grant, Philip A. Romero, and Ophelia S. Venturelli. 2020. “Investigating the dynamics of microbial consortia in spatially structured environments.” Nature Communications 11 (1). https://doi.org/10.1038/s41467-020-16200-0.
Halleran, Andrew D., Emanuel Flores-Bautista, and Richard M. Murray. 2019. “Quantitative characterization of random partitioning in the evolution of plasmid-encoded traits,” March, 594879. https://doi.org/10.1101/594879.
Hogg, David W, Jo Bovy, and Dustin Lang. n.d. “Data analysis recipes: Fitting a model to data *.” http://arxiv.org/abs/1008.4686v1.
Holzer, Matt, Arjen Doelman, and Tasso J. Kaper. 2013. “Existence and stability of traveling pulses in a reaction-diffusion- mechanics system.” 1. Vol. 23. https://doi.org/10.1007/s00332-012-9147-0.
Hsiao, Victoria, Yutaka Hori, Paul WK Rothemund, and Richard M Murray. 2016. “A population‐based temporal logic gate for timing and recording chemical events.” Molecular Systems Biology 12 (5): 869. https://doi.org/10.15252/msb.20156663.
Jain, A., S. Singh, B. Kumar Sarma, and H. Bahadur Singh. 2012. “Microbial consortium-mediated reprogramming of defence network in pea to enhance tolerance against Sclerotinia sclerotiorum.” Journal of Applied Microbiology. https://doi.org/10.1111/j.1365-2672.2011.05220.x.
Jin, Zi Jing, Lian Zhou, Shuang Sun, Ying Cui, Kai Song, Xuehong Zhang, and Ya Wen He. 2020. “Identification of a Strong Quorum Sensing- And Thermo-Regulated Promoter for the Biosynthesis of a New Metabolite Pesticide Phenazine-1-carboxamide in Pseudomonas strain PA1201.” ACS Synthetic Biology 9 (7): 1802–12. https://doi.org/10.1021/acssynbio.0c00161.
Johns, Nathan I., Tomasz Blazejewski, Antonio L. C. Gomes, and Harris H. Wang. 2016. “Principles for designing synthetic microbial communities.” Current Opinion in Microbiology 31: 146–53. https://doi.org/10.1016/j.mib.2016.03.010.
Kaplan, H. B., and E. P. Greenberg. 1985. “Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system.” Journal of Bacteriology 163 (3): 1210–4. https://doi.org/10.1128/jb.163.3.1210-1214.1985.
Kim, Hyun Jung, Wenbin Du, and Rustem F. Ismagilov. 2011. “Complex function by design using spatially pre-structured synthetic microbial communities: Degradation of pentachlorophenol in the presence of Hg(ii).” Integrative Biology 3 (2): 126–33. https://doi.org/10.1039/c0ib00019a.
Kim, Jae Kyoung, Ye Chen, Andrew J. Hirning, Razan N. Alnahhas, Krešimir Josić, and Matthew R. Bennett. 2019. “Long-range temporal coordination of gene expression in synthetic microbial consortia.” Nature Chemical Biology. https://doi.org/10.1038/s41589-019-0372-9.
Kong, Wentao, David R. Meldgin, James J. Collins, and Ting Lu. 2018. “Designing microbial consortia with defined social interactions.” Nature Chemical Biology 14 (8): 821–29. https://doi.org/10.1038/s41589-018-0091-7.
Langebrake, Jessica B., Gabriel E. Dilanji, Stephen J. Hagen, and Patrick De Leenheer. 2014. “Traveling waves in response to a diffusing quorum sensing signal in spatially-extended bacterial colonies.” Journal of Theoretical Biology 363: 53–61. https://doi.org/10.1016/j.jtbi.2014.07.033.
Larkin, Joseph W., Xiaoling Zhai, Kaito Kikuchi, Samuel E. Redford, Arthur Prindle, Jintao Liu, Sacha Greenfield, et al. 2018. “Signal Percolation within a Bacterial Community.” Cell Systems 7 (2): 137–145.e3. https://doi.org/10.1016/j.cels.2018.06.005.
Lithgow, James K., Adam Wilkinson, Andrea Hardman, Belen Rodelas, Florence Wisniewski-Dyé, Paul Williams, and J. Allan Downie. 2000. “The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci.” Molecular Microbiology 37 (1): 81–97. https://doi.org/10.1046/j.1365-2958.2000.01960.x.
Liu, Chenli, Xiongfei Fu, Lizhong Liu, Xiaojing Ren, Carlos K. L. Chau, Sihong Li, Lu Xiang, et al. 2011. “Sequential establishment of stripe patterns in an expanding cell population.” Science 334 (6053): 238–41. https://doi.org/10.1126/science.1209042.
Macia, Javier, Romilde Manzoni, Núria Conde, Arturo Urrios, Eulàlia de Nadal, Ricard Solé, and Francesc Posas. 2016. “Implementation of Complex Biological Logic Circuits Using Spatially Distributed Multicellular Consortia.” PLoS Computational Biology 12 (2). https://doi.org/10.1371/journal.pcbi.1004685.
Mimee, Mark, Alex C. Tucker, Christopher A. Voigt, and Timothy K. Lu. 2015. “Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota.” Cell Systems 1 (1): 62–71. https://doi.org/10.1016/j.cels.2015.06.001.
Noorbakhsh, Javad, David J. Schwab, Allyson E. Sgro, Thomas Gregor, and Pankaj Mehta. 2015. “Modeling oscillations and spiral waves in Dictyostelium populations.” Physical Review E - Statistical, Nonlinear, and Soft Matter Physics 91 (6): 62711. https://doi.org/10.1103/PhysRevE.91.062711.
Nunan, Naoise, Karl Ritz, David Crabb, Kirsty Harris, Keijan Wu, John W. Crawford, and Iain M. Young. 2006. “Quantification of the in situ distribution of soil bacteria by large-scale imaging of thin sections of undisturbed soil.” FEMS Microbiology Ecology 37 (1): 67–77. https://doi.org/10.1111/j.1574-6941.2001.tb00854.x.
Oleinik, O. A., A. N. Kolmogorov, and N. S. Piskunov. 2019. “Studies of the Diffusion with the Increasing Quantity of the Substance; Its Application to a Biological Problem *.” https://doi.org/10.1201/9780367810504-7.
Payne, Stephen, Bochong Li, Yangxiaolu Cao, David Schaeffer, Marc D. Ryser, and Lingchong You. 2013. “Temporal control of self-organized pattern formation without morphogen gradients in bacteria.” Molecular Systems Biology 9 (1): 697. https://doi.org/10.1038/msb.2013.55.
Pearson, James P., Christian Van Delden, and Barbara H. Iglewski. 1999. “Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals.” Journal of Bacteriology 181 (4): 1203–10. https://doi.org/10.1128/jb.181.4.1203-1210.1999.
Potvin-Trottier, Laurent, Nathan D. Lord, Glenn Vinnicombe, and Johan Paulsson. 2016. “Synchronous long-term oscillations in a synthetic gene circuit.” Nature 538 (7626): 514–17. https://doi.org/10.1038/nature19841.
Prindle, Arthur, Jintao Liu, Munehiro Asally, San Ly, Jordi Garcia-Ojalvo, and Gürol M. Süel. 2015. “Ion channels enable electrical communication in bacterial communities.” Nature 527 (7576): 59–63. https://doi.org/10.1038/nature15709.
Prindle, Arthur, Phillip Samayoa, Ivan Razinkov, Tal Danino, Lev S. Tsimring, and Jeff Hasty. 2012. “A sensing array of radically coupled genetic ’biopixels’.” Nature 481 (7379): 39–44. https://doi.org/10.1038/nature10722.
Reátegui, Eduardo, Fatemeh Jalali, Aimal H. Khankhel, Elisabeth Wong, Hansang Cho, Jarone Lee, Charles N. Serhan, Jesmond Dalli, Hunter Elliott, and Daniel Irimia. 2017. “Microscale arrays for the profiling of start and stop signals coordinating human-neutrophil swarming.” Nature Biomedical Engineering 1 (7). https://doi.org/10.1038/s41551-017-0094.
Redfield, Rosemary J. 2002. “Is quorum sensing a side effect of diffusion sensing?” Trends in Microbiology 10 (8): 365–70. https://doi.org/10.1016/S0966-842X(02)02400-9.
Saini, Mukesh, Chung Jen Chiang, Si Yu Li, and Yun Peng Chao. 2016. “Production of biobutanol from cellulose hydrolysate by the Escherichia coli co-culture system.” FEMS Microbiology Letters 363 (4). https://doi.org/10.1093/femsle/fnw008.
Sarma, Birinchi Kumar, Sudheer Kumar Yadav, Surendra Singh, and Harikesh Bahadur Singh. 2015. “Microbial consortium-mediated plant defense against phytopathogens: Readdressing for enhancing efficacy.” Elsevier Ltd. https://doi.org/10.1016/j.soilbio.2015.04.001.
Schaerli, Yolanda, Andreea Munteanu, Magüi Gili, James Cotterell, James Sharpe, and Mark Isalan. 2014. “A unified design space of synthetic stripe-forming networks.” Nature Communications 5: 4905. https://doi.org/10.1038/ncomms5905.
Scott, Spencer R., M. Omar Din, Philip Bittihn, Liyang Xiong, Lev S. Tsimring, and Jeff Hasty. 2017. “A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis.” Nature Microbiology 2 (8): 17083. https://doi.org/10.1038/nmicrobiol.2017.83.
Steidler, Lothar, Pieter Rottiers, and Bernard Coulie. 2009. “Actobiotics™ as a novel method for cytokine delivery: The interleukin-10 case.” In Annals of the New York Academy of Sciences, 1182:135–45. https://doi.org/10.1111/j.1749-6632.2009.05067.x.
Taillefumier, Thibaud, Anna Posfai, Yigal Meir, and Ned S. Wingreen. 2017. “Microbial consortia at steady supply.” eLife 6: 1–65. https://doi.org/10.7554/eLife.22644.
Tayar, Alexandra M., Eyal Karzbrun, Vincent Noireaux, and Roy H. Bar-Ziv. 2015. “Propagating gene expression fronts in a one-dimensional coupled system of artificial cells.” Nature Physics 11 (12): 1037–41. https://doi.org/10.1038/nphys3469.
Tecon, Robin, Ali Ebrahimi, Hannah Kleyer, Shai Erev Levi, and Dani Or. 2018. “Cell-to-cell bacterial interactions promoted by drier conditions on soil surfaces.” Proceedings of the National Academy of Sciences of the United States of America 115 (39): 9791–6. https://doi.org/10.1073/pnas.1808274115.
Temme, Karsten. 2019. “2019-Pivot-Bio-Performance-Report.” Blog.pivotbio.com.
Tsoi, Ryan, Feilun Wu, Carolyn Zhang, Sharon Bewick, David Karig, and Lingchong You. 2018. “Metabolic division of labor in microbial systems.” Proceedings of the National Academy of Sciences of the United States of America 115 (10): 2526–31. https://doi.org/10.1073/pnas.1716888115.
Voigt, Christopher A. 2020. “Synthetic biology 2020–2030: six commercially-available products that are changing our world.” Nature Communications 11 (1): 6379. https://doi.org/10.1038/s41467-020-20122-2.
Watts, Duncan J., and Steven H. Strogatz. 1998. “Collective dynamics of ’small-world9 networks.” Nature 393 (6684): 440–42. https://doi.org/10.1038/30918.
Willits, Tracy. 2020. “Pivot Bio PROVEN™ Creates Sustainably Self-Fertilizing Corn.”
Wolpert, L. 1969. “Positional information and the spatial pattern of cellular differentiation.” Journal of Theoretical Biology. https://doi.org/10.1016/S0022-5193(69)80016-0.
Xue, Xiaoru, Chuan Xue, and Min Tang. 2018. “The role of intracellular signaling in the stripe formation in engineered Escherichia coli populations.” Edited by Kevin Painter. PLoS Computational Biology 14 (6): e1006178. https://doi.org/10.1371/journal.pcbi.1006178.
Yates, Edwin A., Bodo Philipp, Catherine Buckley, Steve Atkinson, Siri Ram Chhabra, R. Elizabeth Sockett, Morris Goldner, et al. 2002. “N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa.” Infection and Immunity 70 (10): 5635–46. https://doi.org/10.1128/IAI.70.10.5635-5646.2002.
Youk, Hyun, and Wendell A. Lim. 2014. “Secreting and sensing the same molecule allows cells to achieve versatile social behaviors.” Science 343 (6171): 1242782–2. https://doi.org/10.1126/science.1242782.
Young, Jonathan W., James C. W. Locke, Alphan Altinok, Nitzan Rosenfeld, Tigran Bacarian, Peter S. Swain, Eric Mjolsness, and Michael B. Elowitz. 2012. “Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy.” Nature Protocols 7 (1): 80–88. https://doi.org/10.1038/nprot.2011.432.
Zhang, Haoran, and Gregory Stephanopoulos. 2016. “Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli.” Biotechnology Journal 11 (7): 981–87. https://doi.org/10.1002/biot.201600013.
Zomorrodi, Ali R., and Daniel Segrè. 2016. “Synthetic Ecology of Microbes: Mathematical Models and Applications.” Journal of Molecular Biology 428 (5): 837–61. https://doi.org/10.1016/j.jmb.2015.10.019.